※ User Guide:
Frequently Asked Questions:
1.
Q: Is GPS 6.0 much better than the old version?
A:
Yes!
2.
Q: How to use GPS 6.0 webserver?
A:
First, you could find the prediction website in "WEB SERVER" page of GPS 6.0. Second, enter protein sequence(s) in fasta format, which starts with a '>' followed with protein/peptide name. Then, select the kinase node(s). The predictor contains 10 S/T groups, 1 Y group and 2 Dual groups for dual-specificity kinases. Just click the "Submit" button, wait a moment, you can get the prediction results of kinase-specific phosphorylation sites on substrate.
3.
Q: How to choose the different prediction modules? A:
We provide 5 modules of prediction for users. You can click "here >>" at "WEB SERVER" page to change the online service mode or just click the following names:
4.
Q: How to read the GPS 6.0 results? A:
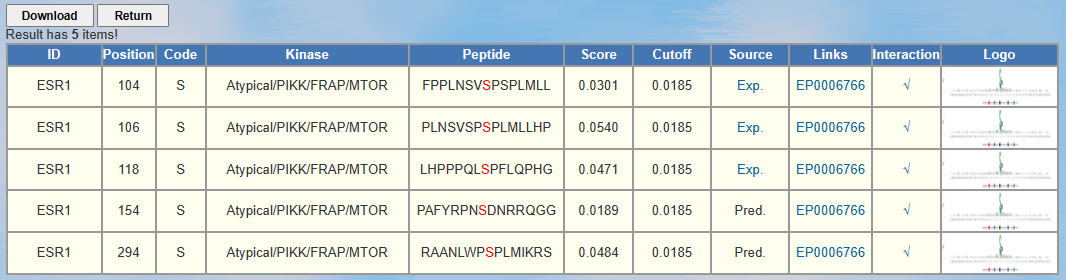
Here we use the human protein ESR1 as the example. After clicking "Submit", the prediction results of MTOR-catalyzed sites with medium threshold are shown as follows:
ID: The name/id of the protein sequence that you input to predict. Position: The position of the site which is predicted to be phosphorylated. Code: The residue which is predicted to be phosphorylated. Kinase: The regulatory kinase which is predicted to phosphorylate the site. Peptide: The predicted phosphopeptide with 7 amino acids upstream and 7 amino acids downstream around the modified residue. Score: The value calculated by GPS 6.0 algorithm to evaluate the potential of phosphorylation. The higher the value, the more potential the residue is phosphorylated. Cutoff: The cutoff value under the threshold. Different threshold means different precision, sensitivity and specificity. Source: Whether this phosphorylation site validated by experiment, "Exp." means YES, while "Pred." means NO. "Exp." links to the source site. Links: The corresponding EPSD page. Interaction: Whether the interaction between this substrate and this kinase validated by BioGrid, "√" that links to the source PubMed page, means YES, while "--" means NO. Logo: The sequence logo of this phosphopeptide. Part 1: Part 2: Part 1: Part 2 : Part 3: 5.
Q: How to choose the cut-off
values and the thresholds? A:
Firstly, we calculated the theoretically maximal
false positive rate (FPR) for each PK cluster.
The three thresholds of GPS 6.0 were decided based
on calculated FPRs.For serine/threonine kinases,
the high, medium and low thresholds were established
with FPRs of 2%, 6% and 10%. And for tyrosine
kinases, the high, medium and low thresholds were
selected with FPRs of 4%, 9% and 15%.
6.
Q: What's the meaning of False
Positive Rate (FPR)? A:
The false positive rate (FPR) is the proportion of negative
sites that are erroneously predicted as positive hits.
Given a data set containing all of non-phosphorylation
sites, the real FPR could be easily computed. However,
precise calculation of FPR is unavailable due to lack
of a "gold-standard" negative data set. Here
we developed a simple and fast method to construct the
near-negative data set and estimate the theoretically
maximal FPRs. Firstly, we calculated the distributions
of amino acids composition in six organisms, including
S. cerevisiae, S. pombe, C. elegans, D. melanogaster, M. musculus,
and H. sapiens. Then we randomly generated
10,000 PSP(30,30) peptides to construct a near-negative
data set based on the real frequencies of twenty amino
acids in eukaryotic proteomes. Although there were a
few sites to be real hits, its proportion would be very
small. The process was repeated twenty times and the
average FPR was calculated by GPS 6.0 as the theoretically
maximal FPR. Also, the negative sites could be randomly
retrieved from eukaryotic proteomes. And the results
from both methods are very similar. 7.
Q: How to use previous versions of GPS?
A:
You can find the previous version of GPS at
DOWNLOAD. Then download
and install the GPS software that you need to your computer.
GPS software is implemented in JAVA and could be installed
on a computer with Windows/Linux/Unix/Mac OS.
And we also wrote a manual for users which included
in the installation package. 8. Q: I was trying to install the software on macbook pro but my installer says the file is damaged. How can I properly install the software in Mac OS? 9. Q: I have a few questions which
are not listed above, how can I contact the authors
of GPS 6.0? A:
Please contact the major author: Miaomiao Chen, Weixhi Zhang, and Dr.
Yu Xue for details. 10. Q:
Can I use GPS 6.0 on different browsers?
A: Yes, we tested our web server on different browsers.
(1) GPS 6.0 has better predictive performance. First, we trained a general model based on 3 types of machine learning algorithms (penalized logistic regression, light gradient boosting machine and deep neural network) using 490,762 non-redundant p-sites. Then, we used transfer learning strategy to construct the kinase-specific models for predicting kinase-specific phosphorylation sites, which improved the predictive accuracy and robustness.
(2) GPS 6.0 deleted some kinases with low confidence of its kinase activity.
(3) The web server of GPS 6.0 can provide more visualizations and is more user-friendly. Users can select different modules to meet different conditions, for example, the id-search module could be selected to perform a task according to the input protein name(s).
(1) GPS Web Server (LightGBM): The default web has fast speed and visualization function. We provide 3D structure, statistics and disorder propensity of protein.
(2) GPS Web Server (Integrated): The deep-learning prediction balances the accuracy with speed.
(3) GPS Web Server of Comprehensive Prediction: The comprehensive prediction has annotations of secondary structure and surface accessibility.
(4) GPS Web Server of Species-specific Prediction: We provide 185 species for species-specific prediction. If you want to focus on certain species, you may choose this one.
(5) GPS Web Server of Prediction by Protein Identifiers : If you want to predict with gene name, protein name or UniProt Accession, please choose this one.
<1>. The table of the GPS 6.0 results
<2>. The visualization of default prediction
Up: The visualization for the positional distribution of the predicted site in protein sequence. By default, the sites with the highest 3 predicted scores are displayed.
Down: The visualization for protein disordered region predicted by IUPred
[PMID: 15955779]. Cutoff = 0.5, if score of prediction > cutoff, the residue is considered in disordered region.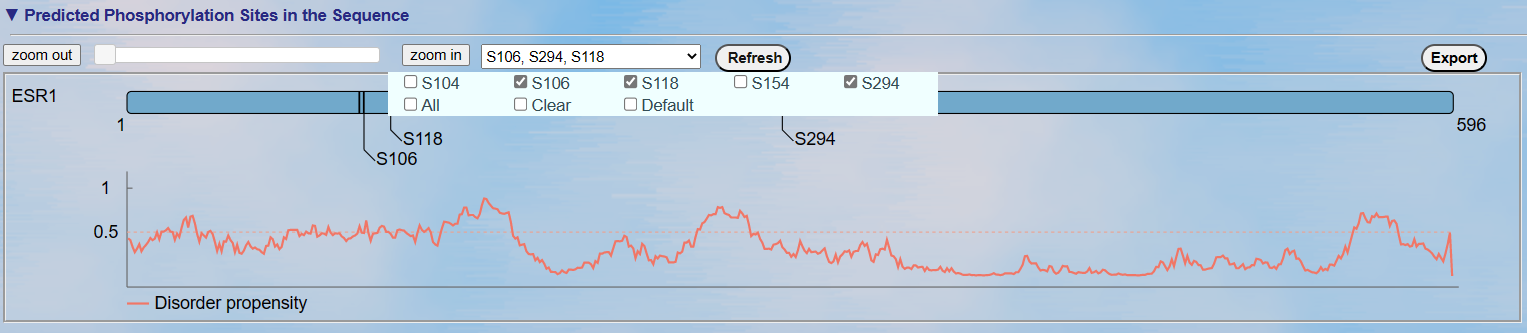
Left: The distribution of S/T/Y sites in kinase families.
The distribution of S/T/Y sites.
The distribution of S/T/Y sites in disordered region.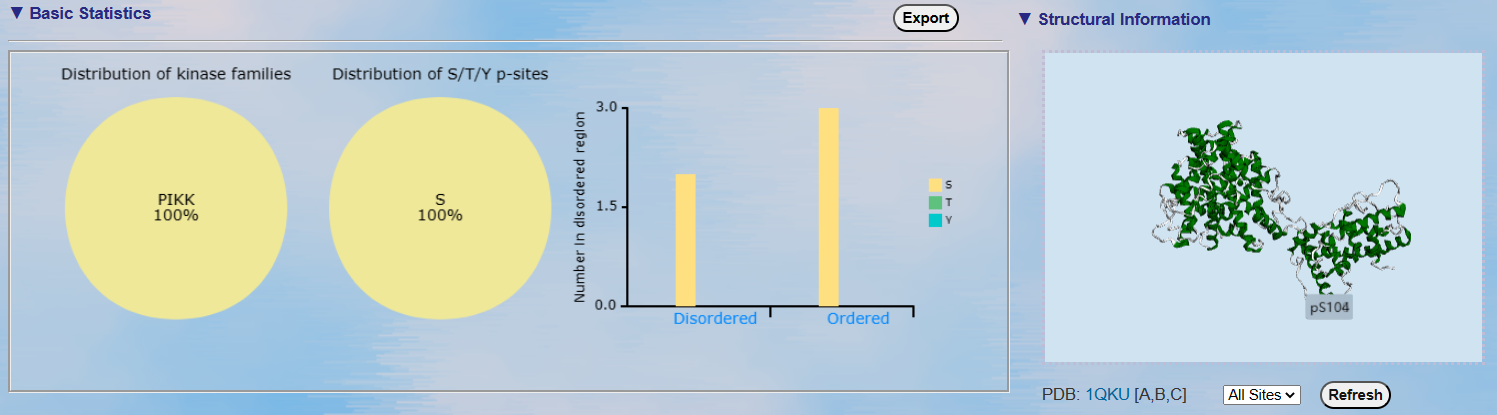
<3>. The visualization of comprehensive prediction
Top: The surface accessibility of amino acids and the protein disordered region were predicted by
NetSurfP ver. 1.1 (PMID: 19646261) and IUPred
(PMID: 15955779), respectively. The cutoff of disordered region prediction = 0.5, if score of prediction > cutoff,
the residue is considered in disordered region. The cutoff of surface accessibility prediction = 0.25, if score of prediction > cutoff, the residue is considered as surface exposed residue.
Bottom: The positions of the predicted phosphorylation sites were visualized in the protein sequence together with the secondary structure predicted by NetSurfP ver. 1.1 (PMID: 19646261).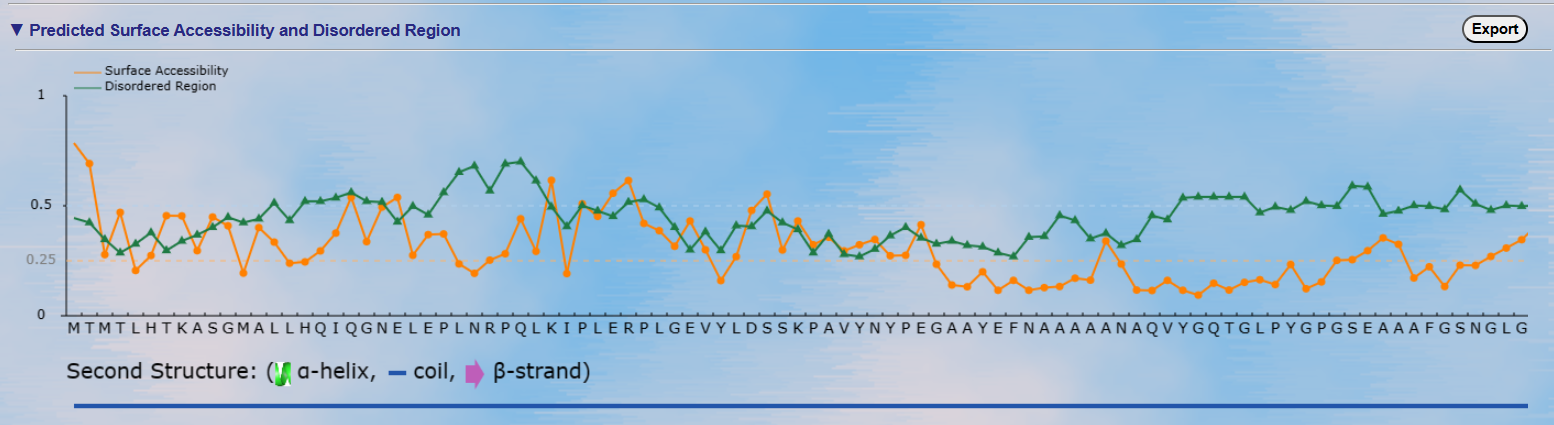
Left: The distribution of S/T/Y sites in kinase families.
Middle left: The distribution of S/T/Y sites.
Middle right: The distribution of S/T/Y sites in secondary structure.
Right: The distribution of S/T/Y sites in disordered region.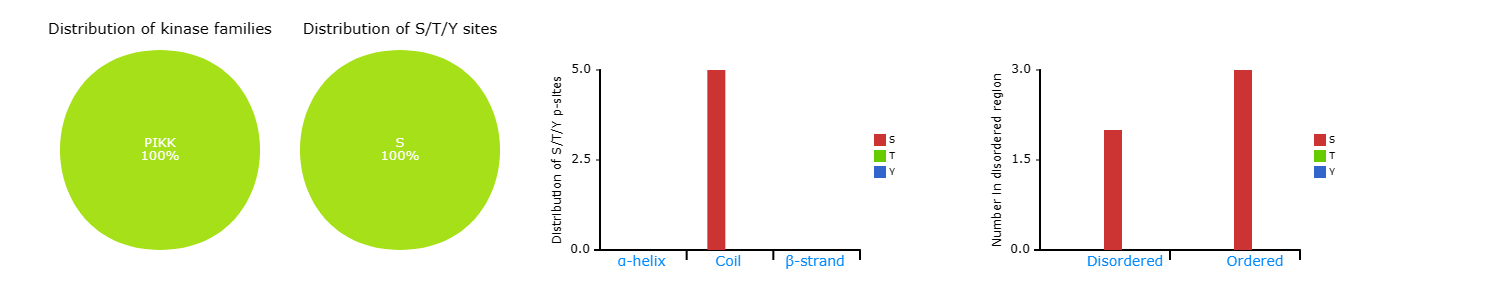
The 3D structure of the substrate labeled with predicted phosphorylation sites.

(1) Open the Preferences. This can be done by either clicking on the System Preferences icon in the Dock or by going to Apple Menu > System Preferences.
(2) Open the Security & Privacy pane by clicking Security & Privacy.
(3) Make sure that the General section of the the Security & Privacy pane is selected. Click the icon labeled Click the lock to prevent further changes.
(4) Enter your username and password into the prompt that appears and click Unlock.
(5) Under the section labeled Allow applications downloaded from, select Anywhere. On the prompt that appears, click Allow From Anywhere.
(6) Exit System Preferences by clicking the red button in the upper left of the window. You should now be able to install applications downloaded from the internet.
OS Version Chrome Firefox Microsoft Edge Safari Linux Ubuntu 18.04 107.0.5304.107 107.0.1 N/A N/A MacOS HighSierra 107.0.5304.107 107.0.1 N/A 16.3 Windows 10 107.0.5304.107 107.0.1 112.0.1722.48 N/A

